Applied Computational Drug Discovery and Design
Our current projects in computer-guided drug discovery focus on the identification and optimisation of bioactive small molecules that
- disrupt protein-protein interactions
- recover the activity of mutated, dysfunctional proteins or stabilise their structure
- are active on drug-resistant microorganisms
- target highly flexible biomacromolecules.
We employ a wide range of computational methods to
- analyse the structure and function of biomacromolecules, in particular also the implications of mutations on protein structure, function, flexibility and ligand binding:
- Protein structure modelling
- Molecular dynamics simulations
- screen large virtual libraries for bioactive synthetic compounds and natural products:
- Docking
- Pharmacophore modelling
- Alignment-based methods, etc.
- identify the likely mode of action (i.e. the macromolecular targets) of small molecules:
- Machine learning
- 2D and 3D similarity-based approaches
- structure-based approaches, etc.
- design innovative ligands based on novel molecular scaffolds:
- De novo design
- elucidate and analyse quantitative structure-activity and structure-property relationships (QSARs/QSPRs):
- Classical QSAR and QSPR models
- Machine learning
- predict the metabolic fate and toxicity of small molecules:
- Machine learning
- Alignment-based and structure-based approaches
- identify compounds prone to trigger false readouts in biological assays:
- Machine learning
- analyse the physicochemical property and scaffold space covered by large molecular libraries, and for diversity analysis and compound selection:
- Clustering, etc.
The molecular libraries that we compiled in our laboratory for virtual screening include:
- >10 billion on-demand compounds that can be obtained at fairly low cost
- >10 million compounds that are readily purchasable from reliable commercial sources
- >250 thousand natural products
- >25 thousand readily purchasable natural products
Selected research outcomes
Selected research outcomes
We successfully applied a pharmacophore-based approach to identify thienoquinolines as a novel class of compounds disrupting the protein-protein interaction of PKCepsilon and RACK2, a result of a long-term collaboration with pharmacologists and chemists. We also identified novel chemical chaperones that correct phenylketonuria in mice with an alignment-based approach. These success stories encouraged us to look more deeply into this interesting topic, and to extend our collaborations with academic partners in Germany, Austria and the UK.
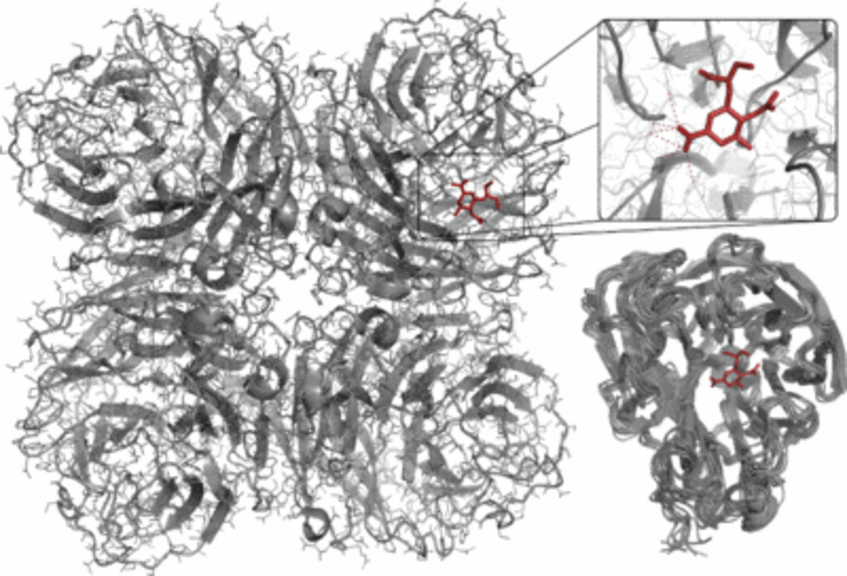
Influenza neuraminidase is the primary target of anti-influenza drugs and is known for its significant conformational flexibility. In recent years influenza strains resistant to marketed drugs have emerged, and hence the development of effective drugs is of utmost importance to public health. Together with researchers at the University of Jena, University of Innsbruck and University of Vienna, we have identified synthetic and natural product inhibitors of influenza neuraminidase, and our lab also contributed to the understanding of the underlying inhibitory mechanisms by observations from molecular dynamics simulations. One of our most recent successes is the discovery of natural products and synthetic molecules inhibiting both viral and bacterial neuraminidases, hence disrupting the lethal synergism between Streptococcus pneumoniae and influenza virus.
We have also found new leads inhibiting the viral coat protein 1 of coxsackievirus B3 and revealed the mechanism of drug resistance caused by mutations of coat protein 1 that are located distant from the ligand (i.e. pocket factor) binding site. Further works include the identification and design of inhibitors for several anti-viral targets, e.g. HIV-1 integrase and reverse transcriptase.
More recently we have engaged in a number of collaborations to identify novel anti-bacterial agents, e.g. N-quinazolinone-4-hydroxy-2-quinolone-3-carboxamides targeting DNA gyrase B, xylose-derived compounds targeting LpxC, and small molecules inhibiting LsrK.
Publications on early drug discovery projects guided by in silico predictions
- Xue, W.; Wang, Y.; Lian, X.; Li, X.; Pang, J.; Kirchmair, J.; Wu, K.; Han, Z.; You, X.; Zhang, H.; Xia, J.; Wu, S., Discovery of N-quinazolinone-4-hydroxy-2-quinolone-3-carboxamides as DNA gyrase B-targeted antibacterial agents. Journal of Enzyme Inhibition and Medicinal Chemistry 2022, 37, 1620-1631.
- Kirchweger, B.; Klein-Junior, L. C.; Pretsch, D.; Chen, Y.; Cretton, S.; Gasper, A. L.; Heyden, Y. V.; Christen, P.; Kirchmair, J.; Henriques, A. T.; Rollinger, J. M., Azepine-indole alkaloids from Psychotria nemorosa modulate 5-HT2A receptors and prevent in vivo protein toxicity in transgenic Caenorhabditis elegans. Frontiers in Neuroscience 2022, 16, 826289.
- Ingwersen, T.; Linnenberg, C.; D’Acunto, E.; Temori, S.; Paolucci, I.; Wasilewski, D.; Mohammadi, B.; Kirchmair, J.; Glen, R. C.; Miranda, E.; Glatzel, M.; Galliciotti, G., G392E neuroserpin causing the dementia FENIB is secreted from cells but is not synaptotoxic. Scientific Reports 2021, 11, 1-13.
- Dreger, A.; Hoff, K.; Agoglitta, O.; Hotop, S. K.; Bronstrup, M.; Heisig, P.; Kirchmair, J.; Holl, R., Antibacterial activity of xylose-derived LpxC inhibitors - Synthesis, biological evaluation and molecular docking studies. Bioorganic Chemistry 2021, 107, 104603.
- Linciano, P.; Cavalloro, V.; Martino, E.; Kirchmair, J.; Listro, R.; Rossi, D.; Collina, S., Tackling antimicrobial resistance with small molecules targeting LsrK: Challenges and opportunities. Journal of Medicinal Chemistry 2020, 63, 15243-15257.
- Xue, W.; Li, X.; Ma, G.; Zhang, H.; Chen, Y.; Kirchmair, J.; Xia, J.; Wu, S., N-thiadiazole-4-hydroxy-2-quinolone-3-carboxamides bearing heteroaromatic rings as novel antibacterial agents: Design, synthesis, biological evaluation and target identification. European Journal of Medicinal Chemistry 2020, 188, 112022.
- Wald, J.; Pasin, M.; Richter, M.; Walther, C.; Mathai, N.; Kirchmair, J.; Makarov, V. A.; Goessweiner-Mohr, N.; Marlovits, T. C.; Zanella, I.; Real-Hohn, A.; Verdaguer, N.; Blaas, D.; Schmidtke, M., Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site. Proceedings of the National Academy of Sciences of the U.S.A. 2019, 1904732116.
- Ehm, P. A. H.; Lange, F.; Hentschel, C.; Jepsen, A.; Bettin, B.; de Bruyn Kops, C.; Kirchmair, J.; Nalaskowski, M.; Jücker, M., Analysis of the FLVR motif of SHIP1 and its relevance to the stability of SH2 containing signaling proteins. Cellular Signalling 2019, 63, 109380.
- Drexel, M.; Kirchmair, J.; Santos-Sierra, S. INH14, a small-molecule urea derivative, inhibits the IKKα/β-dependent TLR inflammatory response. ChemBioChem 2019, 20, 710-717.
- Galster, M.; Löppenberg, M.; Galla, F.; Börgel, F.; Agoglitta, O.; Kirchmair, J.; Holl, R. Phenylethylene glycol-derived LpxC inhibitors with diverse Zn2+-binding groups. Tetrahedron 2018. 75, 486-509.
- Grienke, U.; Maier, C. E.; Kirchmair, K.; Schmidtke, M.; Rollinger, J. M. Discovery of bioactive natural products for the treatment of acute respiratory infections–an integrated approach. Planta Medica 2018, 9/10, 684-695.
- Hoffmann, A.; Richter, M.; von Grafenstein, S.; Walther, E.; Xu, Z.; Schumann, L.; Grienke, U.; Mair, C. E.; Kramer, C.; Rollinger, J. M.; Liedl, K. R.; Schmidtke, M.; Kirchmair, J. Discovery and characterization of diazenylaryl sulfonic acids as inhibitors of viral and bacterial neuraminidases. Frontiers in Microbiology 2017, 8, 205.
- Grienke, U.; Richter, M.; Walther, E.; Hoffmann, A.; Kirchmair, J.; Makarov, V.; Nietzsche, S.; Schmidtke, M.; Rollinger, J. M. Discovery of prenylated flavonoids with dual activity against influenza virus and Streptococcus pneumoniae. Scientific Reports 2016, 27156.
- Walther, E.; Xu, Z.; Richter, M.; Kirchmair, J.; Grienke, U.; Rollinger, J. M.; Krumbholz, A.; Saluz, H. P.; Pfister, W.; Sauerbrei, A.; Schmidtke, M. Dual acting neuraminidase inhibitors open new opportunities to disrupt the lethal synergism between Streptococcus pneumoniae and influenza virus. Frontiers in Microbiology 2016, 7, 357.
- Thelemann, J.; Illarionov, B.; Barylyuk, K.; Geist, J.; Kirchmair, J.; Schneider, P.; Anthore, L.; Root, K.; Trapp, N.; Bacher, A.; Witschel, M.; Zenobi, R.; Fischer, M.; Schneider, G.; Diederich, F. Aryl bis-sulfonamide inhibitors of IspF from Arabidopsis thaliana and Plasmodium falciparum. ChemMedChem 2015, 10, 2090–2098.
- Braun, H.; Kirchmair, J.; Williamson, M. J.; Makarov, V. A.; Riabova, O. B.; Glen, R. C.; Sauerbrei, A.; Schmidtke, M. Molecular mechanism of a specific capsid binder resistance caused by mutations outside the binding pocket. Antiviral Research 2015, 123, 138–145.
- Makarov, V. A.; Braun, H.; Richter, M.; Riabova, O. B.; Kirchmair, J.; Kazakova, E. S.; Seidel, N.; Wutzler, P.; Schmidtke, M. Pyrazolopyrimidines: Potent Inhibitors Targeting the Capsid of Rhino- and Enteroviruses. ChemMedChem 2015, 10, 1629–1634.
- Richter, M.; Schumann, L.; Walther, E.; Hoffmann, A.; Braun, H.; Grienke, U.; Rollinger, J. M.; von Grafenstein, S.; Liedl, K. R.; Kirchmair, J.; Wutzler, P.; Sauerbrei, A.; Schmidtke, M. Complementary assays helping to overcome challenges for identifying neuraminidase inhibitors. Future Virology 2015, 10, 77-88
- Walther, E.; Richter, M.; Xu, Z.; Kramer, C.; von Grafenstein, S.; Kirchmair, J.; Grienke, U.; Rollinger, J. M.; Liedl, K. R.; Slevogt, H.; Sauerbrei, A.; Saluz, H. P.; Pfister, W.; Schmidtke, M. Antipneumococcal activity of neuraminidase inhibiting artocarpin. International Journal of Medical Microbiology 2015, 305, 289–297.
- von Grafenstein, S.; Wallnöfer, H. G.; Kirchmair, J.; Fuchs, J. E.; Huber, R. G.; Spitzer, G.; Schmidtke, M.; Rollinger, J. M.; Liedl, K. R. Interface dynamics explain assembly dependency of influenza neuraminidase catalytic activity. Journal of Biomolecular Structure & Dynamics 2015, 33, 104-120.
- Rechfeld, F.; Gruber, P.; Kirchmair, J. (shared 1st); Boehler, M.; Hauser, N.; Hechenberger, G.; Garczarczyk, D.; Lapa, G. B.; Preobrazhenskaya, M. N.; Goekjian, P.; Langer, T.; Hofmann, J. Thienoquinolines as novel disruptors of the PKCepsilon/RACK2 protein-protein interaction. Journal of Medicinal Chemistry 2014, 57, 3235–3246.
- Grienke, U.; Braun, H.; Seidel, N.; Kirchmair, J.; Richter, M.; Krumbholz, A.; von Grafenstein, S.; Liedl, K. R.; Schmidtke, M.; Rollinger, J. M. Computer-guided approach to access the anti-influenza activity of licorice constituents. Journal of Natural Products 2014, 77, 563–570.
- Xuan, S.; Wang, M.; Kang, H.; Kirchmair, J.; Tan, L.; Yan, A. Support vector machine (SVM) models for predicting inhibitors of the 3’ processing step of HIV-1 integrase. Molecular Informatics 2013, 32, 811-826.
- Santos-Sierra, S.; Kirchmair, J.; Perna, A. M.; Reiß, D.; Kemter, K.; Röschinger, W.; Glossmann, H.; Gersting, S. W.; Muntau, A. C.; Wolber, G.; Lagler, F. Novel pharmacological chaperones that correct phenylketonuria in mice. Human Molecular Genetics 2012, 21, 1877-1887.
- Distinto, S.; Esposito, F.; Kirchmair, J.; Cardia, M. C.; Gaspari, M.; Maccioni, E.; Alcaro, S.; Markt, P.; Wolber, G.; Zinzula, L.; Tramontano, E. Identification of HIV-1 reverse transcriptase dual inhibitors by a combined shape-, 2D-fingerprint- and pharmacophore-based virtual screening approach. European Journal of Medicinal Chemistry 2012, 50, 216–229.
- Grienke, U.; Schmidtke, M.; von Grafenstein, S.; Kirchmair, J.; Liedl, K. R.; Rollinger, J. M. Influenza neuraminidase: A druggable target for natural products. Natural Product Reports 2012, 29, 11-36.
- Rechfeld, F.; Gruber, P.; Hofmann, J.; Kirchmair, J. Modulators of protein-protein interactions – novel approaches in targeting protein kinases and other pharmaceutically relevant biomolecules. Current Topics in Medicinal Chemistry 2011, 11, 1305-1319.
- Gruber, P.; Rechfeld, F.; Kirchmair, J.; Hauser, N.; Boehler, M.; Garczarczyk, D.; Langer, T.; Hofmann, J. Barbituric acid derivative BAS 02104951 inhibits PKCepsilon, PKCeta, PKCepsilon/RACK2 interaction, Elk-1 phosphorylation in HeLa and PKCepsilon and eta translocation in PC3 cells following TPA-induction. Journal of Biochemistry 2011, 149, 331-336.
- Maenz, B.; Goetz, V.; Wunderlich, K.; Eisel, J.; Kirchmair, J.; Stech, J.; Stech, O.; Chase, G.; Frank, R.; Schwemmle, M. Disruption of the viral polymerase complex assembly as a novel approach to attenuate influenza a virus. Journal of Biological Chemistry 2011, 286, 8414-8424.
- Kirchmair, J.; Rollinger, J. M.; Liedl, K. R.; Seidel, N.; Krumbholz, A.; Schmidtke, M. Novel neuraminidase inhibitors: Identification, biological evaluation and investigations of the binding mode. Future Medicinal Chemistry 2011, 3, 437-450.
- Kirchmair, J.; Distinto, S.; Liedl, K. R.; Markt, P.; Rollinger, J. M.; Schuster, D.; Spitzer, G.; Wolber, G. Development of anti-viral agents using molecular modelling and virtual screening techniques. Infectious Disorders – Drug Targets 2011, 11, 64-93.
- Grienke, U.; Schmidtke, M.; Kirchmair, J.; Pfarr, K.; Wutzler, P.; Dürrwald, R.; Wolber, G.; Liedl, K. R.; Stuppner, H.; Rollinger, J. M. Antiviral potential and molecular insight into neuraminidase inhibiting diarylheptanoids from Alpinia katsumadai. Journal of Medicinal Chemistry 2010, 53, 778-786.
- Spitzer, G. M.; Wellenzohn, B.; Markt, P.; Kirchmair, J.; Langer, T.; Liedl, K. R. Hydrogen-bonding patterns of minor groove-binder-DNA complexes reveal criteria for discovery of new scaffolds. Journal of Chemical Information and Modeling 2009, 49, 1063-1069.
- Spitzer, G. M.; Fuchs, J. E.; Markt, P.; Kirchmair, J.; Wellenzohn, B.; Langer, T.; Liedl, K. R. Sequence-specific positions of water molecules at the interface between DNA and minor groove binders. ChemPhysChem 2008, 9, 2766-2771.
